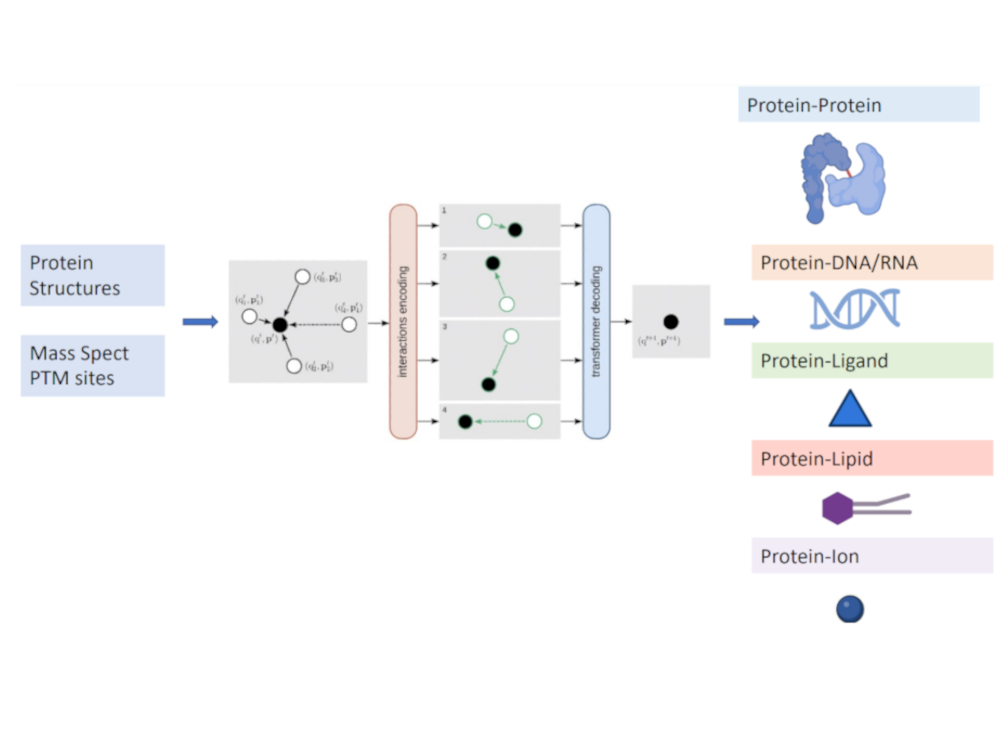
Predict PTMs at inter-molecular interfaces
Identify PTMs at intermolecular interfaces with machine learning methods (manuscript in preparation, to be released in Dec)
Post-translational modifications (PTMs) are the covalent modifications of protein residues. They are widely involved in biological processes such as signal transduction, cell cycle control, and DNA repair. PTMs achieve these functions by modulating the intramolecular and intermolecular interactions. With the rapid development of high-throughput mass spectrometry, a large number of PTM sites were revealed. However, there is a lack of understanding of the structural context of these PTMs. With structure prediction and interface prediction tools, we identified a large number of PTMs at a variety of intermolecular interfaces.
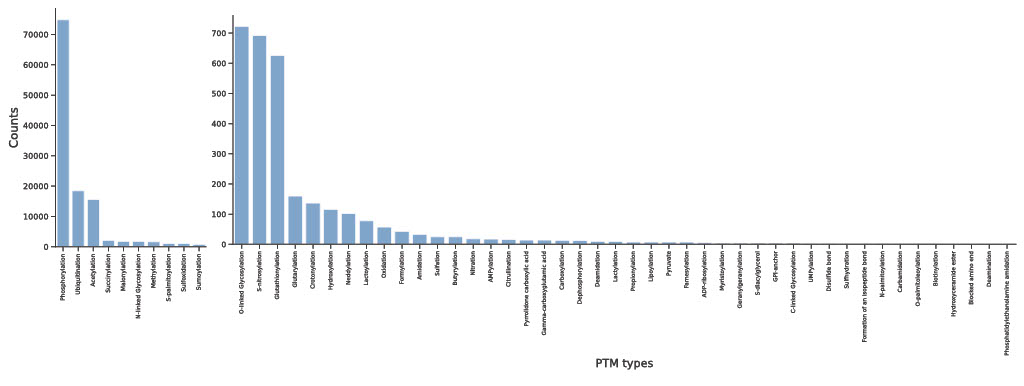
Through our analysis, we found 112,264 AF structures from main organisms with potential PTM sites reported and 1,376,701 PTMs with AF2 structures. Of these, 541,982 PTMs are at the structured region (pLDDT > 70)
The highest number of PTMs reported at these sites is phosphorylation, since phosphorylation is one of the most common PTMs and is most extensively studied PTM (Fig.~
Different PTMs are enriched at various interfaces and potentially regulate distinct biological processes 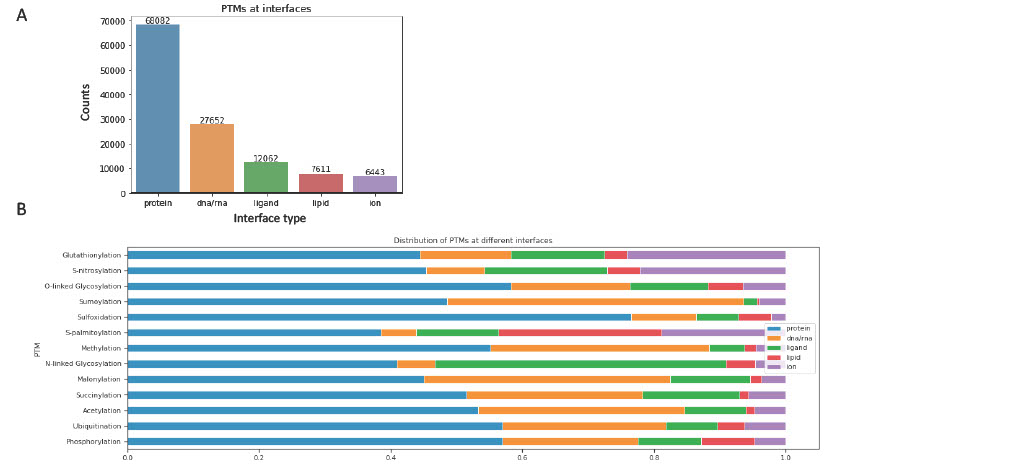
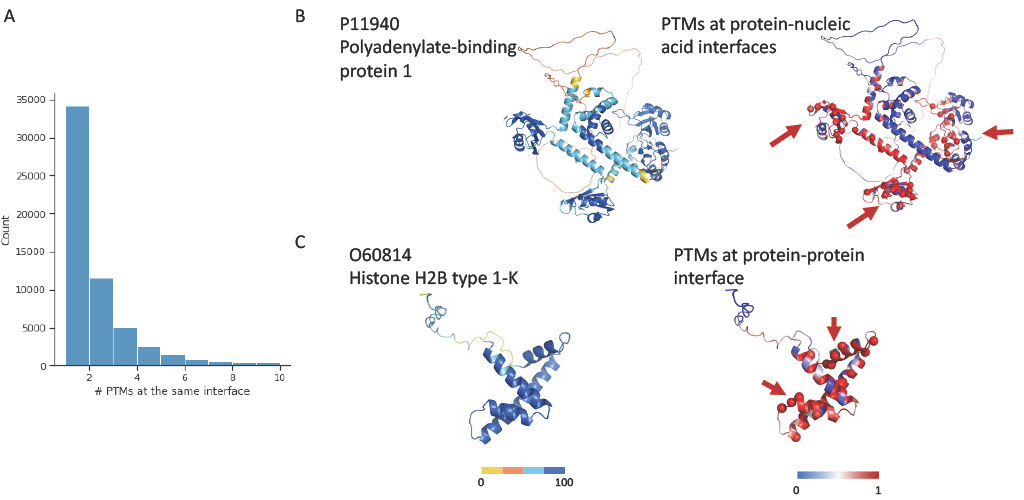
Our analysis revealed that certain interfaces are rich in PTM sites, suggesting that these interfaces might facilitate intricate intermolecular interactions through the coordinated activity of PTM clusters. Among these, 23,923 interfaces have more than one PTM and 3,276 interfaces harbor more than 5 PTMs. This concentration of PTMs within these interface regions may play an essential role in determining the interaction patterns and regulatory mechanisms of the associated proteins.
For example, Polyadenylate-binding protein 1 (PABP1) showcases a PTM hotspot consisting of 44 PTM sites at its protein-nucleic acid interaction interface, with PTMs such as phosphorylation, ubiquitination, acetylation, malonylation, glutathionylation, S-palmitoylation, methylation, and sulfoxidation. PABP1 binds to the poly(A) tail of mRNA and contains multiple PTMs that could influence the pI of PABP and co-regulates its interaction with RNA as well as its interaction with other proteins. In addition, the protein interface of histone H2B type 1-K (H2B1K) also contains a cluster of 16 PTMs sites and 9 PTM types including phosphorylation, acetylation, glutarylation, lactoylation, methylation, succinylation, ubiquitination, sulfoxidation, and sumoylation. The highly dynamic and reversible PTMs of histones as well as the intricate crosstalks between these PTMs are vital for regulating key cellular processes like DNA transcription, replication, and damage repair. This crosstalk is achieved by one PTM modulating the recruitment or activity of enzymes that add/remove another PTM.
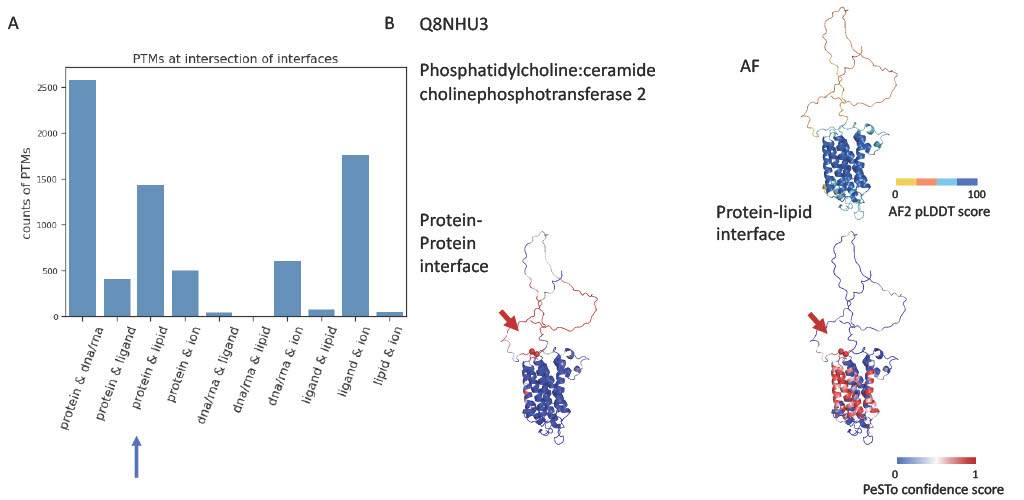
Interestingly, we noticed that certain PTMs exist at the intersection of different types of interfaces. This ‘intersectional’ location of PTMs suggests a versatile role in mediating interactions between a protein and a variety of binding partners. We observed a considerable number of PTMs at the crossover between protein-protein and protein-nucleic acid interfaces as well as between protein-protein and protein-lipid interfaces. The presence of PTMs at such junctures may hint at a multiplex regulation of protein activity and function. An intriguing example of PTMs at interface intersection is Phosphatidylcholine:ceramide cholinephosphotransferase 2 (SMS2), an enzyme that catalyzes the transfer of phosphocholine moiety in sphingomyelin biosynthesis. It is a multi-pass membrane protein, predominantly present in plasma membrane and sometimes present in Golgi apparatus. Palmitoylated on Cys-331, Cys-332 are both at predicted protein interface and lipid interface. Previous research shows that palmitoylation on Cys-331, Cys-332, Cys-343 and Cys-348 plays an important role in plasma membrane localization. The observation that these sites are located at the flexible loop predicted to be both interacting with protein and lipid aligns well with its role in determining membrane localisation and hints that it might also be the binding site of the palmitoylation enzyme.
The intersectional distribution of PTMs across diverse interfaces introduces an intriguing facet of PTM function, suggesting their multifaceted role in coordinating protein interactions and providing a new dimension to our understanding of protein regulation and function.
Given the inherent challenges in synthesizing site-specific PTMs, our understanding of their functional impact is relatively limited. However, extensive research has investigated the effects of mutations and pathogenic natural variants on protein function. These studies provide insights into the biological processes, disease associations, and intermolecular interactions of certain interfaces.Consequently, they can shed light on the potential roles and impacts of PTMs located at these interfaces.
Previous studies have primarily focused on mutations and variants occurring precisely at PTM sites. However, in this study, we adopted a broader perspective by examining mutations and variants found within the same interface as PTMs, regardless of whether they precisely coincide with a PTM site. We extracted mutations and natural variants from UniProt and identified 3350 out of the 58126 total predicted interface with PTMs have a reported mutant with functional info.
6,398 of these mutants have an effect while 717 does not have observed effects (Fig.~
1,433 of these variants are pathogenic while 64 are benign according to dbSNP~\cite{sherry_dbsnp_2001}. Consider the potential connection between disease-relevant variants and PTMs at the same interfaces, it is particularly interesting to examine these PTMs.
Such interfaces could be involved in disease-relevant biological processes that are potentially regulated by these PTMs.